Physicochemical Properties
| Molecular Formula | C40H45N9O9 |
| Molecular Weight | 795.840208768845 |
| Exact Mass | 795.334 |
| CAS # | 2439058-23-6 |
| PubChem CID | 162640960 |
| Appearance | Light yellow to yellow solid |
| LogP | 1.9 |
| Hydrogen Bond Donor Count | 5 |
| Hydrogen Bond Acceptor Count | 12 |
| Rotatable Bond Count | 20 |
| Heavy Atom Count | 58 |
| Complexity | 1410 |
| Defined Atom Stereocenter Count | 0 |
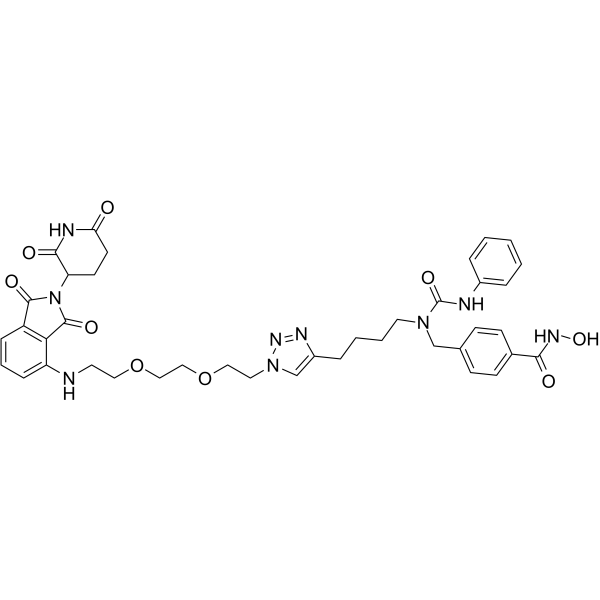
| SMILES | C(NO)(=O)C1=CC=C(CN(CCCCC2=CN(CCOCCOCCNC3=CC=CC4=C3C(=O)N(C3CCC(=O)NC3=O)C4=O)N=N2)C(NC2=CC=CC=C2)=O)C=C1 |
| InChi Key | NLVMRCQAKUCFIA-UHFFFAOYSA-N |
| InChi Code | InChI=1S/C40H45N9O9/c50-34-17-16-33(37(52)43-34)49-38(53)31-10-6-11-32(35(31)39(49)54)41-18-21-57-23-24-58-22-20-48-26-30(44-46-48)9-4-5-19-47(40(55)42-29-7-2-1-3-8-29)25-27-12-14-28(15-13-27)36(51)45-56/h1-3,6-8,10-15,26,33,41,56H,4-5,9,16-25H2,(H,42,55)(H,45,51)(H,43,50,52) |
| Chemical Name | 4-[[4-[1-[2-[2-[2-[[2-(2,6-dioxopiperidin-3-yl)-1,3-dioxoisoindol-4-yl]amino]ethoxy]ethoxy]ethyl]triazol-4-yl]butyl-(phenylcarbamoyl)amino]methyl]-N-hydroxybenzamide |
| HS Tariff Code | 2934.99.9001 |
| Storage |
Powder-20°C 3 years 4°C 2 years In solvent -80°C 6 months -20°C 1 month Note: Please store this product in a sealed and protected environment, avoid exposure to moisture. |
| Shipping Condition | Room temperature (This product is stable at ambient temperature for a few days during ordinary shipping and time spent in Customs) |
Biological Activity
| Targets | HDAC6 |
| ln Vitro | By conjugating a novel HDAC6 inhibitor Nex A with CRBN ligand Poma, a new class of PROTAC degraders of HDAC6 was developed. Among the different HDAC6-targeting PROTACs we developed, NP8 stood out as the most efficient degrader. NP8 effectively induced degradation of HDAC6 at 100 nmol/L in different cell lines, most significantly in multiple myeloma cells. NP8-induced degradation of HDAC6 was a rapid and specific process which required the simultaneous binding of HDAC6 and CRBN and was dependent on proteasome activity. The EGFP-fused HDAC6 was responsive to NP8-induced degradation, suggesting the potential combination of protein degrader with fluorescence techniques to monitor the dynamics of interested proteins. In addition, the comparable inhibitory activity of NP8 and Nex A on multiple myeloma cells implies the future application of novel HDAC6-degradation strategy to treat this notorious disease[1]. |
| Enzyme Assay | Next, to understand the dynamic details of PROTAC-induced HDAC6 degradation, direct visualization was applied for monitoring the process. With the potent and selective HDAC6-targeting degrader NP8, we were able to observe that how the HDAC6 degradation occurred under the treatment of NP8. We fused an enhanced green fluorescence protein (EGFP) to the N-terminal of HDAC6 to track the distribution and dynamics of HDAC6 in cells. The EGFP-HDAC6 showed an exclusively cytoplasmic distribution, consistent with reported HDAC6 subcellular localization (Kawaguchi et al., 2003) (Fig. 2A). When transfected into HeLa, the signals of EGFP-HDAC6 could be attenuated upon the treatment of NP8 (Fig. 2B) and the fusion proteins were indeed degraded under NP8 induction (Fig. 2C). When the signals from several single cells were monitored in time-lapse manner, the significant reduction of mean fluorescence intensities could be observed in a consecutive time window (Fig. 2D). To be noted, the response of EGFP-HDAC6 to NP8 was somehow not as rapid as endogenous HDAC6, possibly due to much higher expression level after transfection. Nonetheless, these data suggested that EGFP-HDAC6 was a responsive indicator for PROTAC-induced degradation and further visualization approaches may also be realized via combination of NP8 and other methods[1]. |
| Cell Assay | To evaluate the degradation capability of our PROTACs for HDAC6 protein, we analyzed the cellular levels of HDAC6 in HeLa cells by Western blot after incubation with four different PROTACs. It was found that all PROTACs can effectively induce HDAC6 degradation after 24 h. Among them, NP8 was the most potent degrader which can significantly reduce the HDAC6 protein level at 100 nmol/L (Fig. 1C). We then went on to evaluate the degradation potential of NP8 in a panel of cell lines from different origins. NP8 consistently induced significant degradation of HDAC6 in all the cell lines we tested, while the multiple myeloma cell line MM.1S exhibited the best sensitivity to NP8 (Fig. S1). The NP8-induced degradation was specific for HDAC6 since the other representative HDAC family members were not affected by NP8 treatment (Figs. 1D and S2). Time-lapse experiment showed that NP8 induced fast and effective degradation of HDAC6 in just 2 h post drug treatment (Fig. 1E). The half degradation concentration (DC50) of NP8 in MM.1S was 3.8 nmol/L (Fig. 1G). The proliferation of MM.1S was inhibited by NP8 in a dose-dependent manner, with significant and comparable level as the parental drug Nex A (Fig. 1H). To further characterize the working mechanisms for NP8, we applied two negative controls, NP8-NC1 and NP8-NC2. NP8-NC1 is an inactive analog of NP8 with a defect in binding CRBN, while NP8-NC2 is the simple conjugate of Poma and linker without Nex A (namely 3 in Scheme 1). Compared to NP8, these two control molecules both failed to induce HDAC6 degradation, implying the essence of simultaneous HDAC6 and CRBN binding for successful degradation (Fig. 1F). In parallel, NP8-induced HDAC6 depletion could be blocked by co-treatment of Nex A or Poma, again indicating NP8 needed binding of CRBN and HDAC6 to fulfil target degradation (Fig. 1F). Additionally, proteasome inhibitor Carfilzomib (CARF) could block the HDAC6 degradation, further demonstrating the dependence on proteasome for PROTAC (Fig. 1F) (Kuhn et al., 2007). Moreover, HDAC6 levels were quickly restored upon NP8 removal (Fig. S3). Collectively, these data demonstrated that the PROTAC NP8 represented a potent and specific degrader of HDAC6. Though small molecule inhibitors of HDAC6 have been studied extensively, there are still some problems remained to be solved, including the potential off-target effects, and the loss of efficacy to gene mutations (Batchu et al., 2016). Additionally, the non-enzymatic functions of HDAC6 cannot be affected by inhibitors. Compared with typical inhibitors, HDAC6-targeting degraders may have advantages over conventional inhibition by small molecules[1]. |
| References |
[1]. Developing potent PROTACs tools for selective degradation of HDAC6 protein [published correction appears in Protein Cell. 2019 Feb 22;:]. Protein Cell. 2019;10(8):606-609. |
Solubility Data
| Solubility (In Vitro) | DMSO :~100 mg/mL (~125.65 mM) |
| Solubility (In Vivo) |
Solubility in Formulation 1: ≥ 2.5 mg/mL (3.14 mM) (saturation unknown) in 10% DMSO + 40% PEG300 +5% Tween-80 + 45% Saline (add these co-solvents sequentially from left to right, and one by one), clear solution. For example, if 1 mL of working solution is to be prepared, you can add 100 μL of 25.0 mg/mL clear DMSO stock solution to 400 μL PEG300 and mix evenly; then add 50 μL Tween-80 + to the above solution and mix evenly; then add 450 μL normal saline to adjust the volume to 1 mL. Preparation of saline: Dissolve 0.9 g of sodium chloride in 100 mL ddH₂ O to obtain a clear solution. (Please use freshly prepared in vivo formulations for optimal results.) |
| Preparing Stock Solutions | 1 mg | 5 mg | 10 mg | |
| 1 mM | 1.2565 mL | 6.2827 mL | 12.5653 mL | |
| 5 mM | 0.2513 mL | 1.2565 mL | 2.5131 mL | |
| 10 mM | 0.1257 mL | 0.6283 mL | 1.2565 mL |