Physicochemical Properties
| Molecular Formula | C17H28CL3N3O |
| Exact Mass | 287.199 |
| Elemental Analysis | C, 51.46; H, 7.11; Cl, 26.80; N, 10.59; O, 4.03 |
| CAS # | 2108631-19-0 |
| Related CAS # | MS023;1831110-54-3;MS023 dihydrochloride;1992047-64-9 |
| PubChem CID | 129626591 |
| Appearance | Typically exists as solid at room temperature |
| Density | 1.1±0.1 g/cm3 |
| Boiling Point | 437.8±45.0 °C at 760 mmHg |
| Flash Point | 218.6±28.7 °C |
| Vapour Pressure | 0.0±1.1 mmHg at 25°C |
| Index of Refraction | 1.567 |
| LogP | 2.3 |
| Hydrogen Bond Donor Count | 5 |
| Hydrogen Bond Acceptor Count | 3 |
| Rotatable Bond Count | 7 |
| Heavy Atom Count | 24 |
| Complexity | 290 |
| Defined Atom Stereocenter Count | 0 |
| InChi Key | VEUUSCXROKBMMJ-UHFFFAOYSA-N |
| InChi Code | InChI=1S/C17H25N3O.3ClH/c1-13(2)21-16-6-4-14(5-7-16)17-11-19-10-15(17)12-20(3)9-8-18;;;/h4-7,10-11,13,19H,8-9,12,18H2,1-3H3;3*1H |
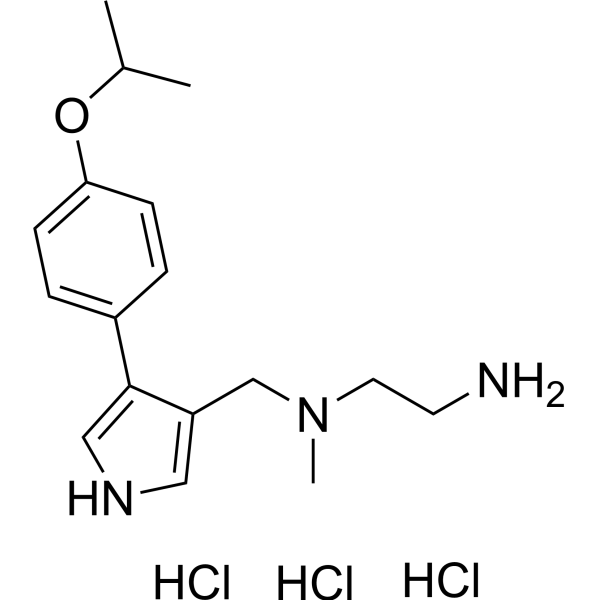
| Chemical Name | N1-((4-(4-isopropoxyphenyl)-1H-pyrrol-3-yl)methyl)-N1-methylethane-1,2-diamine trihydrochloride |
| Synonyms | MS023 trihydrochloride); MS023 triHCl |
| HS Tariff Code | 2934.99.9001 |
| Storage |
Powder-20°C 3 years 4°C 2 years In solvent -80°C 6 months -20°C 1 month |
| Shipping Condition | Room temperature (This product is stable at ambient temperature for a few days during ordinary shipping and time spent in Customs) |
Biological Activity
| Targets | PRMT1 (IC50 = 30 nM); PRMT3 (IC50 = 119 nM); PRMT4 (IC50 = 83 nM); PRMT6 (IC50 = 4 nM); PRMT8 (IC50 = 5 nM) |
| ln Vitro | PRMT1 methyltransferase activity in MCF7 cells is inhibited by MS023 dihydrochloride (1-1000 nM; 48 hours) [1]. PRMT6 methyltransferase activity in HEK293 cells is inhibited by MS023 dihydrochloride (1-1000 nM; 20 hours) [1]. |
| ln Vivo | MS023 dihydrochloride (160 mg/kg, ip) coupled with PKC412 (100 mg/kg, ig) prevents MLL-r timely cycling (ALL) by reducing the support of functional MLL-r ALL starting cells. ) |
| Enzyme Assay |
PRMT biochemical assays[1] A scintillation proximity assay (SPA) was used for assessing the effect of test compounds on inhibiting the methyl transfer reaction catalyzed by PRMTs as described previously.27 In brief, the tritiated S-adenosyl-L-methionine was used as the donor of methyl group. The (3H) methylated biotin labelled peptide was captured in streptavidin/scintillant-coated microplate which brings the incorporated 3H-methyl and the scintillant to close proximity resulting in light emission that is quantified by tracing the radioactivity signal (counts per minute) as measured by a TopCount NXT™ Microplate Scintillation and Luminescence Counter. When necessary, non-tritiated SAM was used to supplement the reactions. The IC50 values were determined under balanced conditions at Km concentrations of both substrate and cofactor by titration of test compounds in the reaction mixture. Cellular PRMT1 assay[1] MCF7 cells were grown in 12-well plates in DMEM supplemented with 10% FBS, penicillin (100 units mL−1) and streptomycin (100 μg mL−1). 40% confluent cells were treated with different concentrations of MS023 and compounds 4 – 6 at indicated concentrations or DMSO control for 48 h. Cells were lysed in 100 μL of total lysis buffer (20 mM Tris-HCl pH 8, 150 mM NaCl, 1 mM EDTA, 10 mM MgCl2, 0.5% TritonX-100, 12.5 U mL−1 benzonase, complete EDTA-free protease inhibitor cocktail). After 3 min incubation at RT, SDS was added to final 1% concentration. Lysates were run on SDS-PAGE and immunoblotting was done as outlined below to determine H4R3me2a, arginine asymmetric dimethylation, arginine symmetric dimethylation and arginine monomethylation in western blot. Cellular PRMT6 assay[1] HEK293 cells were grown in 12-well plates in DMEM supplemented with 10% FBS, penicillin (100 U mL−1) and streptomycin (100 μg mL−1). 50 % confluent cells were transfected with FLAG-tagged PRMT6 or mutant V86K/D88A PRMT6 (1 μg of DNA per well) using jetPRIME® transfection reagent (Polyplus-Transfection), following manufacturer instructions. After 4 h media were removed and cells were treated with MS023 at indicated concentrations or DMSO control. After 20 h, media was removed and cells were lysed in 100 μL of total lysis buffer. |
| Cell Assay |
Western Blot Analysis [1] Cell Types: MCF7 and HEK293 cells Tested Concentrations: 1.4, 4, 12, 37, 111, 333 and 1000 nM Incubation Duration: 48 hrs (hours) for MCF7 cells; 20 hrs (hours) for HEK293 cells Experimental Results:: Treatment potently and concentration-dependently diminished cellular levels of H4R3me2a (IC50=9±0.2 nM). Treatment concentration-dependently diminished the H3R2me2a mark (IC50=56±7 nM). |
| Animal Protocol |
Animal/Disease Models: NOD-scid IL2Rgnull (NSG) mice carrying primary MLL-r ALL cells [2] Doses: 160 mg/kg Route of Administration: intraperitoneal (ip) injection; spread of [2]. Results of 4 weeks of PKC412 (100 mg/kg, ig), MS023 (160 mg/kg, ip) or combination treatment: Combination treatment prolonged the survival of leukemia mice relative to single treatment. |
| References |
[1]. A Potent, Selective, and Cell-Active Inhibitor of Human Type I Protein Arginine Methyltransferases. ACS Chem Biol. 2016 Mar 18;11(3):772-81. [2]. Targeting PRMT1-mediated FLT3 methylation disrupts maintenance of MLL-rearranged acute lymphoblastic leukemia. Blood. 2019 Oct 10;134(15):1257-1268. |
| Additional Infomation |
Protein arginine methyltransferases (PRMTs) play a crucial role in a variety of biological processes. Overexpression of PRMTs has been implicated in various human diseases including cancer. Consequently, selective small-molecule inhibitors of PRMTs have been pursued by both academia and the pharmaceutical industry as chemical tools for testing biological and therapeutic hypotheses. PRMTs are divided into three categories: type I PRMTs which catalyze mono- and asymmetric dimethylation of arginine residues, type II PRMTs which catalyze mono- and symmetric dimethylation of arginine residues, and type III PRMT which catalyzes only monomethylation of arginine residues. Here, we report the discovery of a potent, selective, and cell-active inhibitor of human type I PRMTs, MS023, and characterization of this inhibitor in a battery of biochemical, biophysical, and cellular assays. MS023 displayed high potency for type I PRMTs including PRMT1, -3, -4, -6, and -8 but was completely inactive against type II and type III PRMTs, protein lysine methyltransferases and DNA methyltransferases. A crystal structure of PRMT6 in complex with MS023 revealed that MS023 binds the substrate binding site. MS023 potently decreased cellular levels of histone arginine asymmetric dimethylation. It also reduced global levels of arginine asymmetric dimethylation and concurrently increased levels of arginine monomethylation and symmetric dimethylation in cells. We also developed MS094, a close analog of MS023, which was inactive in biochemical and cellular assays, as a negative control for chemical biology studies. MS023 and MS094 are useful chemical tools for investigating the role of type I PRMTs in health and disease.[1] Relapse remains the main cause of MLL-rearranged (MLL-r) acute lymphoblastic leukemia (ALL) treatment failure resulting from persistence of drug-resistant clones after conventional chemotherapy treatment or targeted therapy. Thus, defining mechanisms underlying MLL-r ALL maintenance is critical for developing effective therapy. PRMT1, which deposits an asymmetric dimethylarginine mark on histone/non-histone proteins, is reportedly overexpressed in various cancers. Here, we demonstrate elevated PRMT1 levels in MLL-r ALL cells and show that inhibition of PRMT1 significantly suppresses leukemic cell growth and survival. Mechanistically, we reveal that PRMT1 methylates Fms-like receptor tyrosine kinase 3 (FLT3) at arginine (R) residues 972 and 973 (R972/973), and its oncogenic function in MLL-r ALL cells is FLT3 methylation dependent. Both biochemistry and computational analysis demonstrate that R972/973 methylation could facilitate recruitment of adaptor proteins to FLT3 in a phospho-tyrosine (Y) residue 969 (Y969) dependent or independent manner. Cells expressing R972/973 methylation-deficient FLT3 exhibited more robust apoptosis and growth inhibition than did Y969 phosphorylation-deficient FLT3-transduced cells. We also show that the capacity of the type I PRMT inhibitor MS023 to inhibit leukemia cell viability parallels baseline FLT3 R972/973 methylation levels. Finally, combining FLT3 tyrosine kinase inhibitor PKC412 with MS023 treatment enhanced elimination of MLL-r ALL cells relative to PKC412 treatment alone in patient-derived mouse xenografts. These results indicate that abolishing FLT3 arginine methylation through PRMT1 inhibition represents a promising strategy to target MLL-r ALL cells.[2] |
Solubility Data
| Solubility (In Vitro) | May dissolve in DMSO (in most cases), if not, try other solvents such as H2O, Ethanol, or DMF with a minute amount of products to avoid loss of samples |
| Solubility (In Vivo) |
Note: Listed below are some common formulations that may be used to formulate products with low water solubility (e.g. < 1 mg/mL), you may test these formulations using a minute amount of products to avoid loss of samples. Injection Formulations (e.g. IP/IV/IM/SC) Injection Formulation 1: DMSO : Tween 80: Saline = 10 : 5 : 85 (i.e. 100 μL DMSO stock solution → 50 μL Tween 80 → 850 μL Saline) *Preparation of saline: Dissolve 0.9 g of sodium chloride in 100 mL ddH ₂ O to obtain a clear solution. Injection Formulation 2: DMSO : PEG300 :Tween 80 : Saline = 10 : 40 : 5 : 45 (i.e. 100 μL DMSO → 400 μLPEG300 → 50 μL Tween 80 → 450 μL Saline) Injection Formulation 3: DMSO : Corn oil = 10 : 90 (i.e. 100 μL DMSO → 900 μL Corn oil) Example: Take the Injection Formulation 3 (DMSO : Corn oil = 10 : 90) as an example, if 1 mL of 2.5 mg/mL working solution is to be prepared, you can take 100 μL 25 mg/mL DMSO stock solution and add to 900 μL corn oil, mix well to obtain a clear or suspension solution (2.5 mg/mL, ready for use in animals). Injection Formulation 4: DMSO : 20% SBE-β-CD in saline = 10 : 90 [i.e. 100 μL DMSO → 900 μL (20% SBE-β-CD in saline)] *Preparation of 20% SBE-β-CD in Saline (4°C,1 week): Dissolve 2 g SBE-β-CD in 10 mL saline to obtain a clear solution. Injection Formulation 5: 2-Hydroxypropyl-β-cyclodextrin : Saline = 50 : 50 (i.e. 500 μL 2-Hydroxypropyl-β-cyclodextrin → 500 μL Saline) Injection Formulation 6: DMSO : PEG300 : castor oil : Saline = 5 : 10 : 20 : 65 (i.e. 50 μL DMSO → 100 μLPEG300 → 200 μL castor oil → 650 μL Saline) Injection Formulation 7: Ethanol : Cremophor : Saline = 10: 10 : 80 (i.e. 100 μL Ethanol → 100 μL Cremophor → 800 μL Saline) Injection Formulation 8: Dissolve in Cremophor/Ethanol (50 : 50), then diluted by Saline Injection Formulation 9: EtOH : Corn oil = 10 : 90 (i.e. 100 μL EtOH → 900 μL Corn oil) Injection Formulation 10: EtOH : PEG300:Tween 80 : Saline = 10 : 40 : 5 : 45 (i.e. 100 μL EtOH → 400 μLPEG300 → 50 μL Tween 80 → 450 μL Saline) Oral Formulations Oral Formulation 1: Suspend in 0.5% CMC Na (carboxymethylcellulose sodium) Oral Formulation 2: Suspend in 0.5% Carboxymethyl cellulose Example: Take the Oral Formulation 1 (Suspend in 0.5% CMC Na) as an example, if 100 mL of 2.5 mg/mL working solution is to be prepared, you can first prepare 0.5% CMC Na solution by measuring 0.5 g CMC Na and dissolve it in 100 mL ddH2O to obtain a clear solution; then add 250 mg of the product to 100 mL 0.5% CMC Na solution, to make the suspension solution (2.5 mg/mL, ready for use in animals). Oral Formulation 3: Dissolved in PEG400 Oral Formulation 4: Suspend in 0.2% Carboxymethyl cellulose Oral Formulation 5: Dissolve in 0.25% Tween 80 and 0.5% Carboxymethyl cellulose Oral Formulation 6: Mixing with food powders Note: Please be aware that the above formulations are for reference only. InvivoChem strongly recommends customers to read literature methods/protocols carefully before determining which formulation you should use for in vivo studies, as different compounds have different solubility properties and have to be formulated differently. (Please use freshly prepared in vivo formulations for optimal results.) |