Physicochemical Properties
| Molecular Formula | C17H20CL2N2O |
| Molecular Weight | 339.259502410889 |
| Exact Mass | 338.0952 |
| CAS # | 2289726-31-2 |
| PubChem CID | 169494473 |
| Appearance | Colorless to light yellow liquid |
| LogP | 4.1 |
| Hydrogen Bond Donor Count | 1 |
| Hydrogen Bond Acceptor Count | 3 |
| Rotatable Bond Count | 7 |
| Heavy Atom Count | 22 |
| Complexity | 319 |
| Defined Atom Stereocenter Count | 0 |
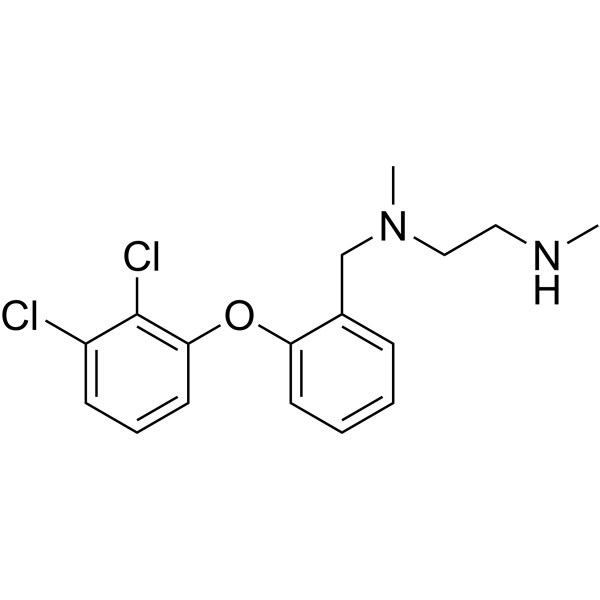
| SMILES | ClC1C(=CC=CC=1OC1C=CC=CC=1CN(C)CCNC)Cl |
| InChi Key | GCURTAFSSBAXNO-UHFFFAOYSA-N |
| InChi Code | InChI=1S/C17H20Cl2N2O/c1-20-10-11-21(2)12-13-6-3-4-8-15(13)22-16-9-5-7-14(18)17(16)19/h3-9,20H,10-12H2,1-2H3 |
| Chemical Name | N'-[[2-(2,3-dichlorophenoxy)phenyl]methyl]-N,N'-dimethylethane-1,2-diamine |
| Synonyms | DCPT1061; DCPT-1061 |
| HS Tariff Code | 2934.99.9001 |
| Storage |
Powder-20°C 3 years 4°C 2 years In solvent -80°C 6 months -20°C 1 month |
| Shipping Condition | Room temperature (This product is stable at ambient temperature for a few days during ordinary shipping and time spent in Customs) |
Biological Activity
| Targets | Protein arginine methyltransferases; PRMT1/6/8 |
| ln Vitro |
In this study, researchers found that PRMT1 expression was remarkably upregulated in tumor tissues and associated with poor pathologic characters and outcomes of ccRCC patients. Furthermore, genetic knockdown and pharmacological inhibition of PRMT1 by a novel potent inhibitor DCPT1061 dramatically induced G1 cell cycle arrest and suppressed ccRCC cell growth. Mechanistically, RNA sequencing and further validation identified Lipocalin2 (LCN2), a secreted glycoprotein implicated in tumorigenesis, as a crucial regulator of ccRCC growth and functional downstream effector of PRMT1. Epigenetic silencing of LCN2 autocrine secretion by PRMT1 deficiency decreased downstream p-AKT, leading to reduced p-RB and cell growth arrest through the neutrophil gelatinase associated lipocalin receptor (NGALR). [2] In this study, researchers found that protein arginine methyltransferase 1 (PRMT1) expression was inversely correlated with the number and activity of CD8+ T cells within melanoma specimen[1]. |
| ln Vivo |
PRMT1 deficiency or inhibition with DCPT1061 significantly restrained refractory melanoma growth and increased intratumoral CD8+ T cells in vivo. Moreover, PRMT1 deletion in melanoma cells facilitated formation of double-stranded RNA derived from endogenous retroviral elements (ERV) and stimulated an intracellular interferon response. Mechanistically, PRMT1 deficiency repressed the expression of DNA methyltransferase 1 (DNMT1) by attenuating modification of H4R3me2a and H3K27ac at enhancer regions of Dnmt1, and DNMT1 downregulation consequently activated ERV transcription and the interferon signaling. Importantly, PRMT1 inhibition with DCPT1061 synergized with PD-1 blockade to suppress tumor progression and increase the proportion of CD8+ T cells as well as IFNγ+CD8+ T cells in vivo. [1] Moreover, PRMT1 inhibition by DCPT1061 not only inhibited tumor growth but also sensitized ccRCC to sunitinib treatment in vivo by attenuating sunitinib-induced upregulation of LCN2-AKT-RB signaling. Conclusion: Taken together, our study revealed a PRMT1-dependent epigenetic mechanism in the control of ccRCC tumor growth and drug resistance, indicating PRMT1 may serve as a promising target for therapeutic intervention in ccRCC patients[2]. |
| Cell Assay | The cultured cells were treated with DCPT1061 or shPRMT1 lentivirus respectively in vitro in 10cm dish plates. 48 h later, cells were collected, and total RNA was isolated. The well-constructed cDNA libraries were then sequenced on the Illumina HiSeq2000 using paired-end methods. After careful quality control of raw reads including trimming adapter sequences and filtering raw reads with low quality, the clean reads were then analyzed through a routine RNA sequencing analysis pipeline. The sequencing reads were first aligned to human hg19 genome by STAR 2.5 (Spliced Transcripts Alignment to a Reference) and then featureCounts software was used to quantify gene expression. Based on these raw counts, differential gene expression analysis was conducted using R/Bioconductor package DESeq2. To define genes differentially expressed in each sample, both fold change of 2 and an adjusted P-value of 0.05 were set as the cut-off value in each case[2]. |
| Animal Protocol |
Mouse tumor model[1] Female 6–8-week-old BALB/c nude mice and female 6- to 8-week-old C57BL/6J wild-type (WT) mice were used. For genetic intervention, 2.5×105 B16-F10 cells for PRMT1 sg1 and CTRL were subcutaneously administrated into WT mice or nude mice. The anti-CD8α antibody (200 μg, intraperitoneal injection) were delivered to mice bearing CTRL or PRMT1 sg1 on day 0, 4, 8, 12. Tumor size and mice body weight were measured every 3 days. When tumors reached 1,000 to 1,500 mm3, mice were sacrificed. For pharmacological intervention, 2.5×105 B16-F10 cells were subcutaneously injected into WT mice or nude mice. Once the tumor volume had reached 50 to 100 mm3, the mice were randomly grouped and intraperitoneal administrated with either vehicle (PBS) or 30 mg/kg DCPT1061 once daily. The anti–PD-1 antibody (150 μg, intraperitoneal injection) were delivered to mice on day 8, 12 alone or partnered with DCPT1061. Tumor size and mice body weight were measured every 3 days. When tumors reached 1,000 to 1,500 mm3, mice were sacrificed. For survival analysis, the mice were considered dead when tumor volume reach to 2,000 mm3. A formula was used to calculate the tumor volume: 1/2 × length × width2. Xenograft model and treatments[2] Around 5 × 106 ccRCC cells were injected subcutaneously into the flank region of six-week-old female nude mice (BALB/c-nu/nu). The PDX was established in six-week-old female SCID mice using the PDX#1002523691 tissue. Tumor volume was measured with a caliper, and the estimated tumor volume = length × width2 /2. When tumors reached approximately 50 mm3, four groups (n = 6) were divided randomly. Treatment of mice was as follows: vehicle control, DCPT1061 (30 mg/kg/day), sunitinib (25 mg/kg/day), and a combination of DCPT1061 (30 mg/kg/day) and sunitinib (25 mg/kg/day). Body weights and tumor volumes were measured every second day. After treatment, mice were sacrificed and the tumors were harvested, weighed, fixed with 4% formaldehyde. |
| References |
[1].PRMT1 is a novel molecular therapeutic target for clear cell renal cell carcinoma. Theranostics. 2021 Mar 12;11(11):5387-5403. [2].PRMT1 inhibition activates the interferon pathway to potentiate antitumor immunity and enhance checkpoint blockade efficacy in melanoma. Cancer Res. 2024 Feb 1;84(3):419-433. |
| Additional Infomation |
Despite the immense success of immune checkpoint blockade (ICB) in cancer treatment, many tumors, including melanoma, exhibit innate or adaptive resistance. Tumor-intrinsic T-cell deficiency and T-cell dysfunction have been identified as essential factors in the emergence of ICB resistance. Here, we found that protein arginine methyltransferase 1 (PRMT1) expression was inversely correlated with the number and activity of CD8+ T cells within melanoma specimen. PRMT1 deficiency or inhibition with DCPT1061 significantly restrained refractory melanoma growth and increased intratumoral CD8+ T cells in vivo. Moreover, PRMT1 deletion in melanoma cells facilitated formation of double-stranded RNA derived from endogenous retroviral elements (ERV) and stimulated an intracellular interferon response. Mechanistically, PRMT1 deficiency repressed the expression of DNA methyltransferase 1 (DNMT1) by attenuating modification of H4R3me2a and H3K27ac at enhancer regions of Dnmt1, and DNMT1 downregulation consequently activated ERV transcription and the interferon signaling. Importantly, PRMT1 inhibition with DCPT1061 synergized with PD-1 blockade to suppress tumor progression and increase the proportion of CD8+ T cells as well as IFNγ+CD8+ T cells in vivo. Together, these results reveal an unrecognized role and mechanism of PRMT1 in regulating antitumor T-cell immunity, suggesting PRMT1 inhibition as a potent strategy to increase the efficacy of ICB.[1] Epigenetic alterations are common events in clear cell renal cell carcinoma (ccRCC), and protein arginine methyltransferase 1 (PRMT1) is an important epigenetic regulator in cancers. However, its role in ccRCC remains unclear. Methods: We investigated PRMT1 expression level and its correlations to clinicopathological factors and prognosis in ccRCC patients based on ccRCC tissue microarrays (TMAs). Genetic knockdown and pharmacological inhibition using a novel PRMT1 inhibitor DCPT1061 were performed to investigate the functional role of PRMT1 in ccRCC proliferation. Besides, we confirmed the antitumor effect of PRMT1 inhibitor DCPT1061 in ccRCC cell-derived tumor xenograft (CDX) models as well as patient-derived tumor xenograft (PDX) models. Results: We found PRMT1 expression was remarkably upregulated in tumor tissues and associated with poor pathologic characters and outcomes of ccRCC patients. Furthermore, genetic knockdown and pharmacological inhibition of PRMT1 by a novel potent inhibitor DCPT1061 dramatically induced G1 cell cycle arrest and suppressed ccRCC cell growth. Mechanistically, RNA sequencing and further validation identified Lipocalin2 (LCN2), a secreted glycoprotein implicated in tumorigenesis, as a crucial regulator of ccRCC growth and functional downstream effector of PRMT1. Epigenetic silencing of LCN2 autocrine secretion by PRMT1 deficiency decreased downstream p-AKT, leading to reduced p-RB and cell growth arrest through the neutrophil gelatinase associated lipocalin receptor (NGALR). Moreover, PRMT1 inhibition by DCPT1061 not only inhibited tumor growth but also sensitized ccRCC to sunitinib treatment in vivo by attenuating sunitinib-induced upregulation of LCN2-AKT-RB signaling. Conclusion: Taken together, our study revealed a PRMT1-dependent epigenetic mechanism in the control of ccRCC tumor growth and drug resistance, indicating PRMT1 may serve as a promising target for therapeutic intervention in ccRCC patients.[2] |
Solubility Data
| Solubility (In Vitro) | DMSO :≥ 250 mg/mL (~736.90 mM) |
| Solubility (In Vivo) |
Note: Listed below are some common formulations that may be used to formulate products with low water solubility (e.g. < 1 mg/mL), you may test these formulations using a minute amount of products to avoid loss of samples. Injection Formulations (e.g. IP/IV/IM/SC) Injection Formulation 1: DMSO : Tween 80: Saline = 10 : 5 : 85 (i.e. 100 μL DMSO stock solution → 50 μL Tween 80 → 850 μL Saline) *Preparation of saline: Dissolve 0.9 g of sodium chloride in 100 mL ddH ₂ O to obtain a clear solution. Injection Formulation 2: DMSO : PEG300 :Tween 80 : Saline = 10 : 40 : 5 : 45 (i.e. 100 μL DMSO → 400 μLPEG300 → 50 μL Tween 80 → 450 μL Saline) Injection Formulation 3: DMSO : Corn oil = 10 : 90 (i.e. 100 μL DMSO → 900 μL Corn oil) Example: Take the Injection Formulation 3 (DMSO : Corn oil = 10 : 90) as an example, if 1 mL of 2.5 mg/mL working solution is to be prepared, you can take 100 μL 25 mg/mL DMSO stock solution and add to 900 μL corn oil, mix well to obtain a clear or suspension solution (2.5 mg/mL, ready for use in animals). Injection Formulation 4: DMSO : 20% SBE-β-CD in saline = 10 : 90 [i.e. 100 μL DMSO → 900 μL (20% SBE-β-CD in saline)] *Preparation of 20% SBE-β-CD in Saline (4°C,1 week): Dissolve 2 g SBE-β-CD in 10 mL saline to obtain a clear solution. Injection Formulation 5: 2-Hydroxypropyl-β-cyclodextrin : Saline = 50 : 50 (i.e. 500 μL 2-Hydroxypropyl-β-cyclodextrin → 500 μL Saline) Injection Formulation 6: DMSO : PEG300 : castor oil : Saline = 5 : 10 : 20 : 65 (i.e. 50 μL DMSO → 100 μLPEG300 → 200 μL castor oil → 650 μL Saline) Injection Formulation 7: Ethanol : Cremophor : Saline = 10: 10 : 80 (i.e. 100 μL Ethanol → 100 μL Cremophor → 800 μL Saline) Injection Formulation 8: Dissolve in Cremophor/Ethanol (50 : 50), then diluted by Saline Injection Formulation 9: EtOH : Corn oil = 10 : 90 (i.e. 100 μL EtOH → 900 μL Corn oil) Injection Formulation 10: EtOH : PEG300:Tween 80 : Saline = 10 : 40 : 5 : 45 (i.e. 100 μL EtOH → 400 μLPEG300 → 50 μL Tween 80 → 450 μL Saline) Oral Formulations Oral Formulation 1: Suspend in 0.5% CMC Na (carboxymethylcellulose sodium) Oral Formulation 2: Suspend in 0.5% Carboxymethyl cellulose Example: Take the Oral Formulation 1 (Suspend in 0.5% CMC Na) as an example, if 100 mL of 2.5 mg/mL working solution is to be prepared, you can first prepare 0.5% CMC Na solution by measuring 0.5 g CMC Na and dissolve it in 100 mL ddH2O to obtain a clear solution; then add 250 mg of the product to 100 mL 0.5% CMC Na solution, to make the suspension solution (2.5 mg/mL, ready for use in animals). Oral Formulation 3: Dissolved in PEG400 Oral Formulation 4: Suspend in 0.2% Carboxymethyl cellulose Oral Formulation 5: Dissolve in 0.25% Tween 80 and 0.5% Carboxymethyl cellulose Oral Formulation 6: Mixing with food powders Note: Please be aware that the above formulations are for reference only. InvivoChem strongly recommends customers to read literature methods/protocols carefully before determining which formulation you should use for in vivo studies, as different compounds have different solubility properties and have to be formulated differently. (Please use freshly prepared in vivo formulations for optimal results.) |
| Preparing Stock Solutions | 1 mg | 5 mg | 10 mg | |
| 1 mM | 2.9476 mL | 14.7380 mL | 29.4759 mL | |
| 5 mM | 0.5895 mL | 2.9476 mL | 5.8952 mL | |
| 10 mM | 0.2948 mL | 1.4738 mL | 2.9476 mL |