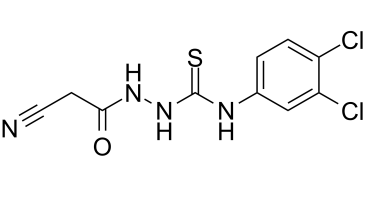
iKIX1 is a novel and potent inhibitor of Pdr1-dependent gene activation with antifungal activity. It is able to resensitize drug-resistant C. glabrata to azole antifungals in vitro and in animal models for disseminated and urinary tract C. glabrata infection, also inhibits the interaction between the KIX domain of the mediator subunit CgGal11A and the activation domain of CgPdr1, the IC50 and Ki values are 190.2 μM and 18 μM, respectively.
Physicochemical Properties
| Molecular Formula | C10H8CL2N4OS |
| Molecular Weight | 303.17 |
| Exact Mass | 301.979 |
| Elemental Analysis | C, 39.62; H, 2.66; Cl, 23.39; N, 18.48; O, 5.28; S, 10.58 |
| CAS # | 656222-54-7 |
| PubChem CID | 2803648 |
| Appearance | Solid powder |
| Density | 1.6±0.1 g/cm3 |
| Index of Refraction | 1.681 |
| LogP | 2.26 |
| Hydrogen Bond Donor Count | 3 |
| Hydrogen Bond Acceptor Count | 3 |
| Rotatable Bond Count | 2 |
| Heavy Atom Count | 18 |
| Complexity | 370 |
| Defined Atom Stereocenter Count | 0 |
| SMILES | C1(C=CC(NC(NNC(CC#N)=O)=S)=CC=1Cl)Cl |
| InChi Key | XKQJVBSDCOCZFV-UHFFFAOYSA-N |
| InChi Code | InChI=1S/C10H8Cl2N4OS/c11-7-2-1-6(5-8(7)12)14-10(18)16-15-9(17)3-4-13/h1-2,5H,3H2,(H,15,17)(H2,14,16,18) |
| Chemical Name | 2-Cyano-acetic acid 2-[[(3,4-dichlorophenyl)amino]thioxomethyl]hydrazide |
| Synonyms | iKIX1; iKIX-1; iKIX 1; |
| HS Tariff Code | 2934.99.9001 |
| Storage |
Powder-20°C 3 years 4°C 2 years In solvent -80°C 6 months -20°C 1 month |
| Shipping Condition | Room temperature (This product is stable at ambient temperature for a few days during ordinary shipping and time spent in Customs) |
Biological Activity
| ln Vitro | In the presence of 5 µM ketoconazole (KET), iKIX1 (10–20 μg/ml) reduces cell proliferation in HepG2 cells in a concentration-dependent manner [1]. With a fitted Kd of 319.7 nM, the FP titration curve illustrates the interaction between the CgGal11A KIX domain and the CgPdr1 AD30. CgPdr1 AD30 and iKIX1 are in competition with an IC50 of 190.2 µM. During in vitro binding experiments, iKIX1 demonstrated an apparent Ki of 18 µM and a Kd of 0.32 µM for the CgPdr1 activation domain (AD) of the CgGal11A KIX domain [1]. In the Sc pdr1Δpdr3Δ strain, which carries plasmid-borne CgPDR1 and 3XPDRE-luciferase, iKIX1 (0-50 µM) dose-responsively suppresses the increase of luciferase activity produced by KET [1]. In Saccharomyces cerevisiae, the recruitment of Gal11/Med15 to Pdr1-regulated target genes was investigated using the chromatin immunoprecipitation (ChIP) assay. Gal11/Med15 is quickly recruited by ketoconazole to the promoters of Pdr1 target genes PDR5 and SNQ2. Ketoconazole-induced Gal11/Med15 recruitment is eliminated by iKIX1, and azole-induced ScPdr1 target gene transcription is significantly suppressed [1]. There is an impact of iKIX1 (20 μM) on C's transcription. glabrata Pdr1-regulated genes involved in drug efflux and MDR (CgCDR1, CgCDR2, and CgYOR1). iKIX1 by alone had no discernible impact on Pdr1 target gene induction. On the other hand, iKIX1 pretreatment permanently and concentration-dependently decreased ketoconazole-induced CgPdr1 upregulation [1]. C was the subject of an RNA sequencing (RNA-Seq) experiment. glabrata strain SFY114 (wild-type PDR1). In both yeast species, azole upregulates Pdr1-dependent genes, including the drug efflux pumps ScPDR5 and CgCDR1i. A number of S genes that are Pdr1-dependent and azole-activated are significantly suppressed when KIX1 binds to azoles. cerevisiae as well as S. glabrata, however in S. cerevisiae, iKIX1 alone impacts distinct gene sets. cerevisiae as well as S. Glabrata. Then, either GAL11/MED15 impacts very distinct genomes in S., or iKIX1 does not dramatically change PDR1 expression. cerevisiae as well as S. Glabrata [1]. Azole effectiveness against CgPDR1 gain-of-function mutants is restored by iKIX1 (0-150 µM). In a concentration-dependent manner, it restores azole sensitivity to PDR1 gain-of-function mutants [1]. |
| References | [1]. Joy L Nishikawa, et al.Inhibiting fungal multidrug resistance by disrupting an activator-Mediator interaction. Nature. 2016 Feb 25;530(7591):485-9. |
Solubility Data
| Solubility (In Vitro) |
DMSO : 61 ~83.33 mg/mL (201.2 ~274.86 mM) Ethanol : 1 mg/mL |
| Solubility (In Vivo) |
Solubility in Formulation 1: 2.08 mg/mL (6.86 mM) in 10% DMSO + 40% PEG300 + 5% Tween80 + 45% Saline (add these co-solvents sequentially from left to right, and one by one), suspension solution; with sonication. For example, if 1 mL of working solution is to be prepared, you can add 100 μL of 20.8 mg/mL clear DMSO stock solution to 400 μL PEG300 and mix evenly; then add 50 μL Tween-80 to the above solution and mix evenly; then add 450 μL normal saline to adjust the volume to 1 mL. Preparation of saline: Dissolve 0.9 g of sodium chloride in 100 mL ddH₂ O to obtain a clear solution. Solubility in Formulation 2: ≥ 2.08 mg/mL (6.86 mM) (saturation unknown) in 10% DMSO + 90% (20% SBE-β-CD in Saline) (add these co-solvents sequentially from left to right, and one by one), clear solution. For example, if 1 mL of working solution is to be prepared, you can add 100 μL of 20.8 mg/mL clear DMSO stock solution to 900 μL of 20% SBE-β-CD physiological saline solution and mix evenly. Preparation of 20% SBE-β-CD in Saline (4°C,1 week): Dissolve 2 g SBE-β-CD in 10 mL saline to obtain a clear solution. Solubility in Formulation 3: ≥ 2.08 mg/mL (6.86 mM) (saturation unknown) in 10% DMSO + 90% Corn Oil (add these co-solvents sequentially from left to right, and one by one), clear solution. For example, if 1 mL of working solution is to be prepared, you can add 100 μL of 20.8 mg/mL clear DMSO stock solution to 900 μL of corn oil and mix evenly. Solubility in Formulation 4: 10% DMSO+40% PEG300+5% Tween-80+45% Saline: 2.08 mg/mL (6.86 mM) (Please use freshly prepared in vivo formulations for optimal results.) |
| Preparing Stock Solutions | 1 mg | 5 mg | 10 mg | |
| 1 mM | 3.2985 mL | 16.4924 mL | 32.9848 mL | |
| 5 mM | 0.6597 mL | 3.2985 mL | 6.5970 mL | |
| 10 mM | 0.3298 mL | 1.6492 mL | 3.2985 mL |