SPHINX31 is a novel, highly potent, selective, and cell bioactive inhibitor of the serine/arginine-rich protein kinase 1 (SRPK1) with IC50 value of 6 nM. Serine/arginine splicing factor 1 (SRSF1) was phosphorylated and SRPK1 activity was inhibited by SPHINX31 treatment. This led to alternative splicing of VEGF-A, which changed its pro-angiogenic isoform to an antiangiogenic one. SPHINX31 raises expression of the anti-angiogenic VEGF-A165b splice variant in retinal pigment epithelial (RPE) cells and inhibits phosphorylation of serine/arginine-rich splicing factor 1 (SRSF1), an SRPK1 substrate, in PC3 cells (EC50 = 360 nM). In a mouse model of choroidal neovascularization, SPHINX31 (2 μg per eye) prevents the growth of blood vessels and the infiltration of macrophages into the eyes. The regulation of VEGF-A alternative splicing to pro-angiogenic isoforms is carried out by serine/arginine-protein kinase 1 (SRPK1), and inhibition of SRPK1 can return the balance between pro- and antiangiogenic isoforms to normal physiological levels.
Physicochemical Properties
| Molecular Formula | C27H24F3N5O2 |
| Molecular Weight | 507.51 |
| Exact Mass | 507.19 |
| Elemental Analysis | C, 63.90; H, 4.77; F, 11.23; N, 13.80; O, 6.30 |
| CAS # | 1818389-84-2 |
| Related CAS # | 1818389-84-2 |
| PubChem CID | 91972002 |
| Appearance | White to off-white solid powder |
| LogP | 4 |
| Hydrogen Bond Donor Count | 1 |
| Hydrogen Bond Acceptor Count | 9 |
| Rotatable Bond Count | 6 |
| Heavy Atom Count | 37 |
| Complexity | 742 |
| Defined Atom Stereocenter Count | 0 |
| InChi Key | VURLRACCOCGFDB-UHFFFAOYSA-N |
| InChi Code | InChI=1S/C27H24F3N5O2/c28-27(29,30)20-4-5-23(35-15-13-34(14-16-35)18-21-3-1-2-10-32-21)22(17-20)33-26(36)25-7-6-24(37-25)19-8-11-31-12-9-19/h1-12,17H,13-16,18H2,(H,33,36) |
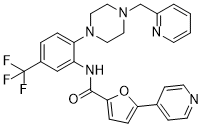
| Chemical Name | 5-pyridin-4-yl-N-[2-[4-(pyridin-2-ylmethyl)piperazin-1-yl]-5-(trifluoromethyl)phenyl]furan-2-carboxamide |
| Synonyms | SPHINX31; SPHINX 31; 1818389-84-2; N-(2-(4-(pyridin-2-ylmethyl)piperazin-1-yl)-5-(trifluoromethyl)phenyl)-5-(pyridin-4-yl)furan-2-carboxamide; 5-pyridin-4-yl-N-[2-[4-(pyridin-2-ylmethyl)piperazin-1-yl]-5-(trifluoromethyl)phenyl]furan-2-carboxamide; 5-(4-pyridinyl)-N-[2-[4-(2-pyridinylmethyl)-1-piperazinyl]-5-(trifluoromethyl)phenyl]-2-furancarboxamide; N-{2-[4-(pyridin-2-ylmethyl)piperazin-1-yl]-5-(trifluoromethyl)phenyl}-5-(pyridin-4-yl)furan-2-carboxamide; SPHINX31?; SPHINX 31;SPHINX-31; SPHINX-31 |
| HS Tariff Code | 2934.99.9001 |
| Storage |
Powder-20°C 3 years 4°C 2 years In solvent -80°C 6 months -20°C 1 month |
| Shipping Condition | Room temperature (This product is stable at ambient temperature for a few days during ordinary shipping and time spent in Customs) |
Biological Activity
| Targets | SRPK1 (IC50 = 5.9 nM); VEGF-A165a |
| ln Vitro |
SPHINX31 was identified as a type 1 kinase inhibitor (ATP competitive) by kinase assays. In PC3 prostate cancer cells, SPHINX31 treatment inhibits SRSF1 phosphorylation at 300 nM. According to mouse liver microsome metabolic stability, SPHINX31 exhibited a medium clearance with a T1/2 of 95.79 min[1]. Leukemic cell differentiation and cell cycle arrest result from SPHINX31-mediated SRPK1 inhibition[2]. Kinase assays showed that SPHINX31 was a type 1 kinase inhibitor (ATP competitive, Supporting Information Figure 2). A radiolabeled ATP competition assay was carried out against 50 kinases, which have been selected by the International Centre for Kinase Profiling as being representative of the human kinome, including SRPK1. This demonstrated 96% inhibition of SRPK1 activity at 1 μM by SPHINX31, but no other kinase was significantly inhibited in that panel (Figure 4b). To determine whether SPHINX31 inhibits SRPK1 activity in cells, PC3 prostate cancer cells (previously been shown to have high SRPK1 mediated SRSF1 phosphorylation (18)) were treated with SPHINX31. This resulted in inhibition of SRSF1 phosphorylation at an inhibitor concentration of 300 nM (Figure 5a,b). Quantitative analysis of the Western blot bands revealed an EC50 of about 360 nM (Figure 5b). The effect on downstream splicing activity was also investigated in retinal pigmented epithelials (RPE, a major source of VEGF in the retina). The compounds dose-dependently switched splicing from VEGF-A165a to VEGF-A165b in RPE cells (Figure 5c).[1] |
| ln Vivo |
SPHINX31 has the potential to enter the eye. In a mouse model, SPHINX31 inhibits choroidal neovascularization in a dose-dependent manner. SPHINX31 prevents macrophage infiltration and blood vessel growth[1]. Immunocompromised mice receiving transplants of MLL-rearranged AML cells have longer survival times when treated with SPHINX31[2]. To investigate the therapeutic potential of SRPK1 inhibition in AML in vivo, we determined the circulating concentration of SPHINX31 after i.p. injection. Injection of 0.8 mg/kg SPHINX31 (i.p.) into DBA2J mice resulted in a concentration of 0.225 ± 0.036 µM in plasma after 24 h. We therefore xenotransplanted RAIL mice with MOLM-13, THP-1 cells or first passage patient-derived AMLs and treated these from day 8 with 0.8 or 2.0 mg/kg of SPHINX31 or vehicle intraperitoneally, for 6 doses over 2 weeks. This led to a significant reduction in leukemic cell growth and a dose-dependent prolongation of survival of mice given MOLM-13, THP-1 and patient-derived MLL-X AMLs (Fig. 1j, k, Supplementary Fig. 5a–i) while the same were not observed with MLL-WT AMLs (Supplementary Fig. 6a–f). These data demonstrate that SRPK1 is a therapeutic vulnerability in MLL-rearranged AMLs.[2] |
| Enzyme Assay |
In Vitro Kinase Screening Assay [1] Candidate compounds were tested for SRPK1 inhibition using a Kinase-Glo assay. A reaction buffer containing 200 mM Tris pH 7.5, 100 mM MgCl2 and 0.1 mg/ml BSA was added to 43 µM SRSF1Arg-Ser (RS) peptide (NH2- RSPSYGRSRSRSRSRSRSRSRSNSRSRSY-OH) and 0.1 µg of purified SRPK1. Candidate compounds were serially diluted from 10 µM to 0.001 nM and added to the reaction mixture, wells with omitted SRPK1 kinase and omitted compounds were also added as controls. All wells contained 1% DMSO. 1 µM ATP was added, wells minus ATP were used as 9 background controls. The plate was then incubated at 30oC for 10 minutes. An equal volume of Kinase-Glo (25 µL) was added to each well and the plate read for luminescence using a Fluostar Optima. Radioactive kinase assays were carried out by the MRC Dundee Kinase Centre. Kinase binding assay was carried out by Kinomescan, Discoverex, at 1 µM. Lack of interference with binding to the SRPK1 substrate was carried out using a dose response curve from 0.5-30 µM. Kinome analysis[2] Kinase binding assay for 489 kinases was carried out by KINOMEscan, DiscoverX, at 1 µM SPHINX31. To identify potential inhibition of kinases other than SRPK1, a truncated version of SRPK1 was used in the screen, which does not contain part of the loop that SPHINX31 binds to, so the SRPK1 activity will not show positivity in this assay. The percent inhibition of kinase-substrate interaction is determined and the red spots correspond to the kinases where there is more 50% inhibition. Radioactive kinase assays were carried out by the MRC Dundee Kinase Centre for SRPK1, SRPK2, CLK1 and CLK2 from 10 µM to 0.0003 µM SPHINX31 with ATP at the Km for each kinase. SPHINX31 SRSF1 phosphorylation[2] 1 × 106 cells/ml, unless otherwise stated, were treated with SPHINX31 at 1% DMSO for 48 h then lysed in buffer containing 50 mM Hepes, 150 mM NaCl, 0.5% Triton-X100, 1 mM EDTA, 1 mM PMSF, 10 mM Na3VO4, 10 mM NaF and protease inhibitor cocktail. 50 µg protein was separated on SDS-PAGE gels and immunoblotted with anti-SRSF1, anti-pSRSF and anti-ACTIN . Isothermal Titration Calorimetry (ITC) The ITC experiment was performed at 30 ˚C using Nano-ITC. The protein in 50mM HEPES, pH 7.5, 500mM NaCl, 0.5mM TCEP, 5% glycerol at 50 µM was titrated into 6 µM SPHINX31. The heated of binding were analyzed, and the data were fitted to an independent binding model using NanoAnalyze program, from which the thermodynamic parameters were calculated (∆G = ∆H - T∆S = -RTlnKB, where ∆G, ∆H and ∆S are the changes in free energy, enthalpy and entropy of binding, respectively). Thermodynamic parameters and binding constants are given in Supplemental Table 2. |
| Cell Assay |
Pharmacological inhibitor treatments. [1] Cells at ~70% confluence were serum starved for 24 h and treated with inhibitors at concentration noted. To determine pSRSF1 levels, cells were pre-treated with SPHINX31 11 for 1 hr then treated with TNF α (50 ng/ml) for 30 mins. For VEGF immunoblots in PC-3 cells, cells were treated with inhibitors for 24 h, washed off and lysed 24 h after washing. Flow cytometry analyses of AML cells[2] Cells were transduced with gRNA vectors or treated with SPHINX31 and stained at the indicated time points with anti-mouse CD11b PE/Cy5 (Biolegend, cat. no. 101210) and anti-human CD11b PE or anti-human CD13 FITC. Data were analyzed by using LSRFortessa and FlowJo. Apoptosis levels were measured in human and/or mouse AML cells transduced with dual gRNA vectors (against SRPK1 and 3’ BCL2 enhancer) and/or treated with 1 or 3 μM SPHINX31 at indicated time points, by using Annexin V. Data were analyzed by using LSRFortessa instruments. Cell cycle stages were measured in human and/or mouse AML cells transduced with dual gRNA vectors against SRPK1 and/or treated with 1 or 3 μM SPHINX31 at indicated time points, using Propidium Iodide. Data were analyzed using LSRFortessa instruments. Drug and proliferation assays[2] All suspension cells were plated (96-well) in triplicate at 5000–10,000 cells per well and treated for 72 h with vehicle or the indicated concentrations of SPHINX31 (0.04-50 μM), Cytarabine (0.075–40 μM), Daunorubicin (0.075–80 μM) and indicated IC20 doses of iBET-151 (0.04–25 μM). On day 3, plates were measured (for treatments with Cytarabine and Daunorubicin) using CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay in order to calculate the relative cell proliferation. Regarding the treatment with SPHINX31, an equal volume for all wells was split-back with fresh media and compound, such that the resulting cell density in each well matched the initial seeding density. Plates were measured on day 6 using CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay in order to calculate the relative cell proliferation. All the compounds were dissolved in DMSO. For synergy studies between SPHINX31 and iBET, THP-1 cells were seeded in 96-well plates at 10,000 cells per well and treated with SPHINX31 (dose range of 0.039–5 μM) and iBET (dose range of 9.8–312.5 nM) in an 8 by 6 matrix. Each treatment was carried out in triplicate. Cells were treated for 72 h, and cell viability was determined using CellTiter 96 AQueous Non-Radioactive Cell Proliferation Assay in order to calculate the relative cell proliferation. Cell viability for each treatment was normalized against the DMSO control group. A Bliss independence model was employed to evaluate combination effects and calculate the Bliss independence score3. All the compounds were dissolved in DMSO. Adult primary leukemia and cord blood sample drug and proliferation assays[2] All human AML and cord blood samples were obtained with informed consent under local ethical approval (REC 07-MRE05-44). Primary human AML cells or cord-blood-derived CD34+ cells were tested for colony-forming efficiency in StemMACS HSC-CFU semi-solid medium (Miltenyi Biotec) in the presence of the indicated concentration of SPHINX31 or DMSO. Colonies were counted by microscopy 11–12 days (AML cells) or 12–14 days (CD34+ cells) after plating. Western blot analysis[2] Cells were treated with indicated concentrations of SPHINX31 or transduced with dual/single lentiviral gRNA or an empty vector and selected with 1.0 μg ml−1 puromycin for 3 days starting from day 2 post-transduction. The transduced cells were further cultured for 2 days before lysis. Cell pellets were resuspended in whole cell lysis buffer (50 mM Tris-HCl, pH = 8, 450 mM NaCl, 0.1% NP-40, 1 mM EDTA), supplemented with 1 mM DTT, protease inhibitors (Sigma), and phosphatase inhibitors. Protein concentrations were assessed by Bradford assay and an equal amount of protein was loaded per track. Prior to loading, the samples were supplemented with SDS-PAGE sample buffer and DTT was added to each sample. 10–40 μg of protein was separated on SDS-PAGE gels, and blotted onto polyvinylidene difluoride membranes. |
| Animal Protocol |
DBA2J mice 0.8 mg/kg i.p. Generation of PDX models[2] Six- to ten-week-old NSG female mice were injected with 106 patient-derived AML cells by intravenous injection. Indicated doses of SPHINX31 or vehicle were delivered to the mice via intraperitoneal injection (IP) on day 10 post-transplant, triweekly for two weeks (6 treatments). Indicated doses of vehicle or SPHINX31 were delivered to the mice via intraperitoneal injection (IP) on day 10 post-transplant. SPHINX31 was dissolved in 20%(w/v) 2-hydroxypropyl-beta-cyclodextrin vehicle. At day 10 post-transplant, tumor burdens of animals were detected using IVIS Lumina II (Caliper) with Living Image version 4.3.1 software. Briefly, 100 μl of 30 mg/ml D-luciferin was injected into each animal intraperitoneally. Ten min after injection, the animals were maintained under general anesthesia by isoflurane and put into the IVIS chamber for imaging. The detected tumor burdens were measured and quantified by the same software. Diseased mice were identified by qualified animal technicians from the Sanger mouse facility. All animal studies were carried out in accordance with the Animals (Scientific Procedures) Act 1986, Amendment Regulations (2012) UK under project license PBF095404. Randomization and blinding were not applied.[2] Whole-body bioluminescent imaging[2] For in vivo experiments, MOLM-13, THP-1 and HEL cells expressing Cas9 were first transduced with a firefly luciferase-expressing plasmid. After propagation, the cells were transduced with a dual lentiviral gRNA vector expressing either empty or SRPK1 gRNA (day 0) and selected with puromycin from day 2 to day 5. At day 5 post-transduction, the cells were suspended in fresh medium without puromycin. At day 6, 1 × 105 cells were transplanted into a Rag2−/− Il2rg−/− mouse by tail-vein injection. For the in vivo drug experiments related to Fig. 1 and Supplementary Figure 4, MOLM-13 and THP-1 cells were transduced with a firefly luciferase-expressing plasmid. 1 × 105 cells were transplanted into a Rag2−/− Il2rg−/− mouse by tail-vein injection. Indicated doses of SPHINX31 or vehicle were delivered to the mice via intraperitoneal injection (IP) on day 10 post-transplant, triweekly for total two weeks (6 treatments). For the in vivo drug experiments related to Fig. 4 and Supplementary Figure 5, THP-1 and HEL cells were transduced with a firefly luciferase– expressing plasmid. 1 × 105 cells were transplanted into a Rag2−/− Il2rg−/− mouse by tail-vein injection. Indicated doses of vehicle, SPHINX31 and/or iBET-151 were delivered to the mice via intraperitoneal injection (IP) from day 10 post-transplantation. Both SPHINX31 and iBET-151 were dissolved in 20%(w/v) 2-hydroxyproply beta-cyclodextrin vehicle (Sigma, H107).[2] At day 10 post-transplant, the tumor burdens of the animals were detected using IVIS Lumina II with Living Image version 4.3.1 software. Briefly, 100 μl of 30 mg/ml D-luciferin (BioVision) was injected into the animals intraperitoneally. Ten min after injection, the animals were maintained in general anesthesia by isoflurane and put into the IVIS chamber for imaging. The detected tumor burdens were measured and quantified by the same software. Diseased mice were assessed blindly by qualified animal technicians from the Sanger mouse facility. All animal studies were carried out in accordance with the Animals (Scientific Procedures) Act 1986, Amendment Regulations (2012) UK under project license PBF095404. Randomization and blinding were not applied.[2] SPHINX31 pharmacokinetics[2] Three Dba/2J mice were given i.p. injections of 0.8 mg/kg SPHINX31 and sacrificed after 24 h when blood was taken by cardiac puncture into EDTA tubes. Plasma was isolated by centrifugation, and an equal volume (100 µl) acetonitrile added. An internal standard of 100 µg/ml of a related compound (compound 3 from Batson et al) was added to samples to account for any loss of material during preparation. The solutions were centrifuged for 15 min at 4 °C and the supernatant taken for analysis. Solutions were evaporated at 37 °C for eight hours and resuspended in 30 µl acetonitrile ready for analysis by LC MS, using a Waters 2795 HPLC system. Detection was achieved by positive ion electrospray (ESI + ) mass spectrometry using a Waters Micromass ZQ spectrometer in single ion monitoring (SIM) mode, at 352 m/z units ([M+H]+). Chromatography (flow rate 1 mL·min−1) was achieved using a Phenomenex Kinetex column (2.6 μ, C18, 100 Å, 4.6 × 50 mm) equipped with a Phenomenex Security Guard precolumn (Luna C5 300 Å). Peaks occurring at these times in the SIM chromatograms per compound were integrated using Water MassLynx software. The chromatograms produced clear peaks at the expected molecular weights. The integrated area under the peaks and read from a standard curve led to quantification of the circulating concentration of SPHINX31.[2] Electroretinography ERG recordings were taken according to ISCEV guidelines using the Phoenix Ganzfeld ERG system. 8 week old female C57/Bl6 mice were treated with a single topical application of 0.2 µg compound 12, 24 h before ERG recordings and dark adapted for at least 12 h and maintained in complete darkness before ERG. Mice were anaesthetized with an intraperitoneal injection of a mixture of 50 mg/kg ketamine and 0.5 mg/kg medetomidine. The pupils were dilated with 5% phenylephrine hydrochloride and 1% tropicamide and kept hydrated with viscotears. Reference electrodes were placed by the tail and scalp and the eye was positioned in contact with the Ganzfeld corneal contact electrode using Labscribe2 ERG and Ueye Cockpit software. Scotopic ERG recordings were taken at 0, 0.02, 0.12, 3.76, 30, 120 and 1920 cd.m.s-2 and photopic recordings at 120 and 1920 cd.m.s-2 alternating in order between right and left eyes to control for any differences. |
| References |
[1]. ACS Chem Biol . 2017 Mar 17;12(3):825-832. [2]. Nat Commun . 2018 Dec 19;9(1):5378. |
| Additional Infomation |
Serine/arginine-protein kinase 1 (SRPK1) regulates alternative splicing of VEGF-A to pro-angiogenic isoforms and SRPK1 inhibition can restore the balance of pro/antiangiogenic isoforms to normal physiological levels. The lack of potency and selectivity of available compounds has limited development of SRPK1 inhibitors, with the control of alternative splicing by splicing factor-specific kinases yet to be translated. We present here compounds that occupy a binding pocket created by the unique helical insert of SRPK1, and trigger a backbone flip in the hinge region, that results in potent (<10 nM) and selective inhibition of SRPK1 kinase activity. Treatment with these inhibitors inhibited SRPK1 activity and phosphorylation of serine/arginine splicing factor 1 (SRSF1), resulting in alternative splicing of VEGF-A from pro-angiogenic to antiangiogenic isoforms. This property resulted in potent inhibition of blood vessel growth in models of choroidal angiogenesis in vivo. This work identifies tool compounds for splice isoform selective targeting of pro-angiogenic VEGF, which may lead to new therapeutic strategies for a diversity of diseases where dysfunctional splicing drives disease development.[1] We recently identified the splicing kinase gene SRPK1 as a genetic vulnerability of acute myeloid leukemia (AML). Here, we show that genetic or pharmacological inhibition of SRPK1 leads to cell cycle arrest, leukemic cell differentiation and prolonged survival of mice transplanted with MLL-rearranged AML. RNA-seq analysis demonstrates that SRPK1 inhibition leads to altered isoform levels of many genes including several with established roles in leukemogenesis such as MYB, BRD4 and MED24. We focus on BRD4 as its main isoforms have distinct molecular properties and find that SRPK1 inhibition produces a significant switch from the short to the long isoform at the mRNA and protein levels. This was associated with BRD4 eviction from genomic loci involved in leukemogenesis including BCL2 and MYC. We go on to show that this switch mediates at least part of the anti-leukemic effects of SRPK1 inhibition. Our findings reveal that SRPK1 represents a plausible new therapeutic target against AML.[2] |
Solubility Data
| Solubility (In Vitro) |
DMSO: 17.3~25 mg/mL (49.3~34.2 mM) Ethanol: ~13 mg/mL (~25.6 mM) |
| Solubility (In Vivo) |
Solubility in Formulation 1: ≥ 2 mg/mL (3.94 mM) (saturation unknown) in 10% DMSO + 40% PEG300 + 5% Tween80 + 45% Saline (add these co-solvents sequentially from left to right, and one by one), clear solution. For example, if 1 mL of working solution is to be prepared, you can add 100 μL of 20.0 mg/mL clear DMSO stock solution to 400 μL PEG300 and mix evenly; then add 50 μL Tween-80 to the above solution and mix evenly; then add 450 μL normal saline to adjust the volume to 1 mL. Preparation of saline: Dissolve 0.9 g of sodium chloride in 100 mL ddH₂ O to obtain a clear solution. Solubility in Formulation 2: ≥ 2 mg/mL (3.94 mM) (saturation unknown) in 10% DMSO + 90% Corn Oil (add these co-solvents sequentially from left to right, and one by one), clear solution. For example, if 1 mL of working solution is to be prepared, you can add 100 μL of 20.0 mg/mL clear DMSO stock solution to 900 μL of corn oil and mix evenly. (Please use freshly prepared in vivo formulations for optimal results.) |
| Preparing Stock Solutions | 1 mg | 5 mg | 10 mg | |
| 1 mM | 1.9704 mL | 9.8520 mL | 19.7040 mL | |
| 5 mM | 0.3941 mL | 1.9704 mL | 3.9408 mL | |
| 10 mM | 0.1970 mL | 0.9852 mL | 1.9704 mL |